Are COVID-19 Vaccines Going To Cause Infertility?
The short version: Someone has claimed that the COVID-19 vaccines are going to cause infertility because of a shared amino acid sequence in the spike protein of SARS-CoV-2 and a placental protein, which will make the immune system attack both as it can’t tell the difference. The truth? This sequence is too short for the immune system to meaningfully confuse it with placental proteins. It’s sort of like saying that you are going to be confused with a criminal because you wear a commonly sold red bracelet that was also found on the criminal. It’s not realistic. If this were true, we would also expect COVID-19 to cause early pregnancy loss a significant amount of the time. The evidence available to us does not support that this is the case. There is no reasonable basis to believe that vaccines against COVID-19/SARS-CoV-2 will affect fertility.
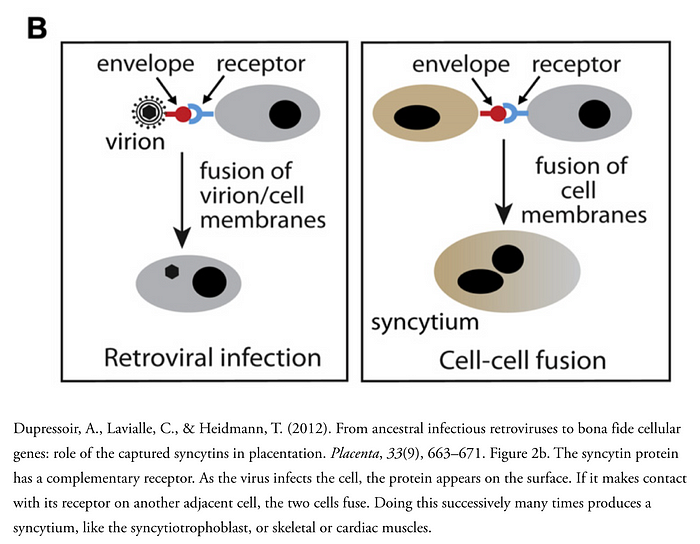
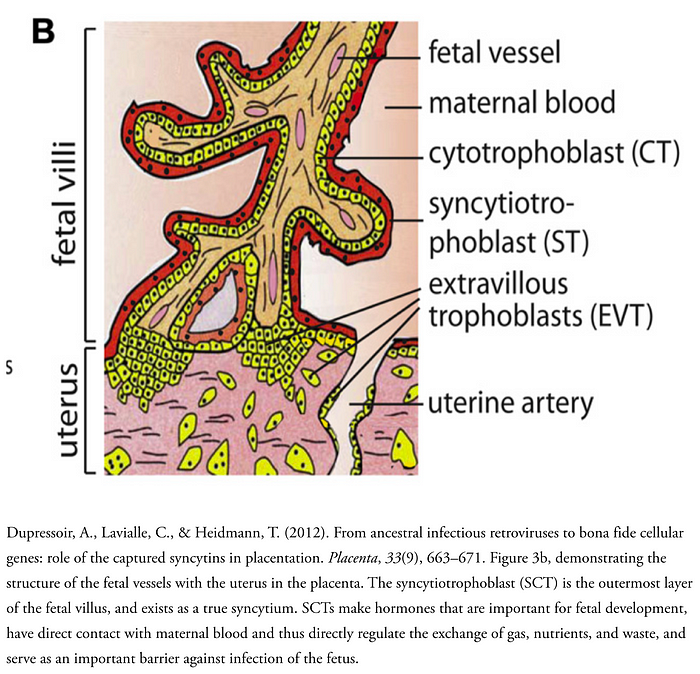
Did you know that the reason that eutherians (placental mammals) have a placenta is because of several retrovirus infections? They gave us proteins called syncytins, which form a structure called a syncytium or polykaryon (literally “many kernels,” where kernel is an old word for “nucleus”) if you’re fancy (you can see how that works on the diagram to the left). A syncytium is essentially one giant cell that contains the nuclei of many cells. Your muscle cells are syncytia (that’s the plural for syncytium), for example, and syncytia are common cytopathic effects one can observe when cell cultures are infected with some viruses. Genetic studies of mice in which syncytins are knocked out (mutations are introduced so that no functional protein is generated) show that syncytins are critical for the formation of a placenta, and in their absence it fails to form. In humans, there are two proteins responsible for this: syncytin-1 (present in every layer of the trophoblast, the outer layer of cells in the blastocyst, a very early point in the development of a mammal appearing between days 5 and 9) and syncytin-2 (found in cytotrophoblast cells). The placenta is required for pregnancy, and thus tampering with syncytins can result in infertility. The placenta is a temporary and autonomous organ made by the fetus (not the mother) which helps to establish a blood supply within the wall of the uterus so that the developing human can exchange nutrients and waste with the mother. In addition, the placenta helps to mediate immunological tolerance between the mother and the fetus. Each is not-self compared with the other, so the placenta forms a barrier between the two so that neither’s immune system can respond to the other.
The placenta has a layer called the syncytiotrophoblast (SCT) which implants into the uterine wall and helps to establish a fetal blood supply, and is in direct contact with the maternal circulation. This is a true syncytium and it has multiple critical supportive functions, including the production of hormones that help to regulate fetal development, the regulation of gas, nutrient, and waste exchange between the fetus and mother, and it serves as a direct barrier against infection. In addition to the SCT, there are so-called giant cells formed from cytotrophoblast that invade into the uterine wall and remodel the vasculature to enhance blood supply to the fetus later in pregnancy. And all this because of a few viruses!
Some have argued that COVID-19 vaccines will cause infertility because of antibodies directed against the spike (S) protein will cross-react with syncytin proteins because of an underlying sequence homology. Let’s talk about whether this claim holds water.
As we’ve established, the SCT supports the overall structure of the placental villi and has many important roles in supporting a pregnancy. SCT cells do indeed express syncytin-1, and an immune response directed against SCT cells can tragically result in early pregnancy loss. People with certain autoimmune diseases like antiphospholipid syndrome (aPS) and systemic lupus erythematosus are (SLE) are at much higher risk for pregnancy loss, which is thought to occur from an inappropriate immune response directed against the trophoblast cells. So that much of it makes sense- if you have an immune response against antigens of the trophoblast, a successful pregnancy is going to be very difficult (pregnancy involves very complex balancing acts from the immune system of mother and fetus. Creasy and Resnik’s Maternal-Fetal Medicine 7th Edition, Chapter 6 Immunology of Pregnancy has very comprehensive coverage of the subject).
The thing is, there is already excellent reason to doubt this is a reasonable claim, and that’s because of COVID-19. While COVID-19 seems to be a more severe disease in pregnancy, there is no evidence right now of an increase in early pregnancy losses, as might be expected if there were an aberrant immune response directed against placental antigens. As Cosma et al write:
Severe acute respiratory syndrome coronavirus 2 infection during the first trimester of pregnancy does not seem to predispose to early pregnancy loss; its cumulative incidence did not differ between women with spontaneous abortion and women with ongoing pregnancy.
Rotshenker-Otsinka et al also add:
The COVID-19 pandemic environment does not seem to affect early first-trimester miscarriage rates in asymptomatic patients.
And there’s even a recent cohort study by Adhikari et al:
In this cohort study of 252 SARS-CoV-2–positive and 3122 negative pregnant women tested in outpatient and inpatient settings at a large county medical center, adverse pregnancy outcomes were similar, and neonatal infection occurred in 3% of infants, predominantly among infants born to asymptomatic or mildly symptomatic women. Placental abnormalities were not associated with disease severity, and the rate of hospitalization was similar to rates among nonpregnant women.
If you look, you will be able to find case reports discussing pregnancy losses in patients with COVID-19, but case reports cannot tell us whether the two events are related, merely that they co-occurred. Though we should not take absence of evidence as evidence of absence, right now there isn’t good data to suggest that having COVID-19 significantly raises the risk of spontaneous abortion, suggesting that at a minimum (factoring in how long the pandemic has been going), if such occurrences are related, they are rare.
Immune responses in COVID-19 are directed against many antigens, including the spike protein, and the intensity of the immune response is far greater than what can be expected from a COVID-19 vaccine. There is also good news in that it seems that vertical transmission of COVID-19 from mother-to-child or mother-to-fetus appears to be very rare, even though the placenta does express ACE2, the receptor for SARS-CoV-2, and it will be important to continue monitoring patients who acquire the virus via vertical transmission to determine if any anomalies result.
In addition to this, the argument is weakened by several other lines of evidence. Firstly, for vaccines containing the receptor-binding domain only, there should not be any expected sequence homology between syncytins and the spike protein, because the receptor-binding domain is not the region of the protein which mediates viral fusion (rather, it is a region between the S1 and S2 regions of the protein, and it requires enzymatic processing to expose this site so that fusion can occur). But, rather than tell you what’s expected, it can be demonstrated that this concern is unreasonable. The Basic Local Alignment Search Tool (BLAST) allows you to input sequences and compare them to known reference sequences to see if there is identity (homology). If you take the sequence of the spike protein (this is the reference sequence) of SARS-CoV-2, there is nothing in reference human genomes that aligns with it:

But if we remove the restriction to humans:

The output of a BLAST search for the spike glycoprotein of SARS-CoV-2. Note that no human sequences align, but there is exceptional homology with many other SARS-CoV-2 isolates.
Addendum
Some additional details have come to light regarding this claim, and they merit an update to the post.
Some additional background on using BLAST searches for these questions is warranted. If very small homologies exist, they will not be detected by the BLAST algorithm (hence the qualifier in the search output of “significant similarity”). There is a sequence of 5 amino acids (there is a divergence for one of them, or else it would be 6) which is shared between the spike protein and the syncytin-1 protein:

I’ll just point out that the same process that generates this result shows that there is a stretch of 9 amino acids with 66% sequence identity between syncytin-1 and SARS-CoV-2 spike, which somehow the author of this claim ignored, even though this would represent a (still untenable but nonetheless) greater risk for autoimmune disease than the short stretch of sequences present there. I mention this to point out the competence of the individual behind the claim only.
This gets missed on BLAST because it does not represent a significant homology, and similarly, it’s not going to be a concern for your immune system. Here’s why: For any kind of molecular mimicry reaction to be possible, we should expect at a minimum that there is a shared epitope. This begs the question of what kind of epitope and how large. T cells recognize linear epitopes, which means you can go just off of the sequence to see if a homology exists. B cells on the other hand recognize native proteins, which are folded and thus recognize conformational epitopes, which may put amino acids next to each other that are far apart on the sequence. It is probably sufficient to look at linear epitopes only because of the nature of linked recognition. Most B cell lineages require T cell help to respond to antigens, and linked recognition ensures that B and T cells respond to the same antigens. There are two exceptions to this. The first is so-called T-independent antigens, of which there are 2 classes:
- T-1 independent (TI-1): These are molecules that induce broad B cell activation that is not based on their ability to recognize antigen. Instead, they generally stimulate B cells to divide extensively, which is known as polyclonal activation. At low concentrations of a TI-1 may falsely demonstrate enrichment of a specific B cell clone.
- T-2 independent (TI-2): Antigens that can cross-link a B cell receptor (immunoglobulin) to activate B cells without the need for T cell help. This mostly applies to carbohydrate antigens which have many repeating units of a sugar sequence.
The other exception are certain so-called extrafollicular B cells. These are special B cell populations that make antibody in the absence of T cell help, often to polysaccharide (carbohydrate i.e. sugar) antigens, and some also make antibodies that have weak affinity for many different antigens, meant mainly as a way for the immune system to buy time early on in the immune response to develop a mature antibody response.
As spike is a protein antigen, it does not meet these concerns (though the protein is heavily glycosylated, these sugars are derived from our own cells and thus should not be able to induce an immune response; if you’re wondering about ABO blood group antigens, we are sensitized to those by our microbiota). Thus, it is prudent to consider T cell epitopes only for the time being. A classic example of molecular mimicry that produces autoimmunity comes from M protein from group A streptococci and rheumatic fever. The M protein is similar to our cardiac myosin, and shares multiple T cell epitopes:

This gives you some idea of the requirements needed for shared T cell epitopes to produce molecular mimicry. However, this table perhaps understates the complexity of the process involved. Let’s suppose I hypothetically found a stretch of 12 shared amino acids between syncytin-1 and the spike protein (I didn’t). Does this represent a legitimate risk for immunologic infertility? The answer is, probably not. The table above shows that you generally need multiple shared T cell epitopes rather than just one. Furthermore, the nature of antigen presentation means this finding alone is insufficient, because how the protein is processed ultimately determines whether or not it forms a T cell epitope, and this is based on a number of factors not limited to the sequence.
The current rule of thumb is for there to be a legitimate molecular mimicry concern as far as B cell epitopes:
usually requires extensive sequence homology, e.g. >35% identity in >50–80 aa peptidic sequences (conformation!) and a linkage of the cross-reacting B-cell epitope to a dominant T helper epitope (foreign). It is facilitated by a strong concomitant activation of innate immunity (danger signal!) [62].
BLAST does not allow for considerations of how proteins fold, and thus cannot model B cell epitopes effectively. A rigorous approach to the question of whether or not molecular mimicry were possible here is actually a fairly advanced bioinformatics project that might merit a publication because one would have to be able to model how the proteins folded in space to see if conformational epitopes were shared (in the case of B cells), where the endogenous antigen presentation machinery is likely to make cuts in processing the spike protein antigen, determine which epitopes would be immunodominant, compare this to known HLA alleles to see if there are specific binding clefts that fit putatively cross-reactive sequences very well to see if a genetic susceptibility exists (assuming a cross-reactive sequence is identified in the first place), and then you would eventually need in vivo validation that this is legitimate (but that comes after all the very heavy bioinformatics work).
So, why did I go with a BLAST search, knowing these deficiencies? Was I being willfully dishonest? Well, no. BLAST will miss very short identities, like the 8–12 amino acids needed to share a T cell epitope. However, if BLAST did identify a significant sequence identity, that could be cause for concern, and thus I felt it was a reasonable shortcut rather than undertaking the complex bioinformatics project I described above, taken together with the available epidemiologic data. I will add though that if a competent bioinformatician wants to undertake the previously aforementioned project, I would be happy to update this post with their findings and credit them fully. The presence of a shared (putative- as it’s not known that this sequence could even end up being presented by antigen-presentation machinery without much more complex analysis) T cell epitope alone does not allow for autoimmune disease- a litany of other conditions must be fulfilled before this can be seriously considered (we still have an extensive suite of immunologic tolerance mechanisms). As the example above shows, multiple T cell epitopes generally have to be shared. Additionally, even in spite of that, rheumatic fever is still a rare consequence of group A streptococcal infection. Furthermore, as immunologist Dr. Croxford notes in his excellent thread on the subject, you can find multiple short sequences that align with the spike protein that are utterly irrelevant.
Lastly, one may wonder, how did this short stretch of amino acids end up in the spike protein? Is it proof that SARS-CoV-2 is a bioweapon? No. Unequivocally, absolutely no. The answer is much more basic than that: evolution. The spike protein evolved to allow SARS-CoV-2 to fuse its membrane with a target cell’s. Syncytins evolved to do the same (but they’re from a different virus). But some sequences will be selected for by evolutionary pressures that enhance transmission. If a particular sequence is selected for because it is good at mediating fusion, this will show up across multiple lifeforms (this is known as convergent evolution). Both syncytin and spike are class I fusion proteins.
In short, the claim that the vaccines for COVID-19/SARS-CoV-2 will cause infertility because of some kind of crossreactive immune response between the spike protein antigens and syncytin proteins is baseless. You should get these vaccines without fear of what they may do for your fertility.
References
- Adhikari, E. H., Moreno, W., Zofkie, A. C., MacDonald, L., McIntire, D. D., Collins, R. R. J., & Spong, C. Y. (2020). Pregnancy outcomes among women with and without severe acute respiratory syndrome Coronavirus 2 infection. JAMA Network Open, 3(11), e2029256
- Cosma, S., Carosso, A. R., Cusato, J., Borella, F., Carosso, M., Bovetti, M., … Benedetto, C. (2020). Coronavirus disease 2019 and first-trimester spontaneous abortion: a case-control study of 225 pregnant patients. American Journal of Obstetrics and Gynecology. doi:10.1016/j.ajog.2020.10.005
- Creasy, R. K., Resnik, R., Iams, J. D., Lockwood, C. J., Moore, T., & Greene, M. F. (2013). Creasy and resnik’s maternal-fetal medicine: Principles and practice (7th ed.). London, England: W B Saunders.
- Cunningham MW. 2019. Molecular mimicry, autoimmunity, and infection: The cross-reactive antigens of group A streptococci and their sequelae. Microbiol Spectr. 7(4). doi:10.1128/microbiolspec.GPP3–0045–2018. http://dx.doi.org/10.1128/microbiolspec.GPP3-0045-2018.
- Dupressoir, A., Lavialle, C., & Heidmann, T. (2012). From ancestral infectious retroviruses to bona fide cellular genes: role of the captured syncytins in placentation. Placenta, 33(9), 663–671.
- Ewald DR, Sumner SCJ. 2018. Human microbiota, blood group antigens, and disease. Wiley Interdiscip Rev Syst Biol Med. 10(3):e1413.
- Faé KC, da Silva DD, Oshiro SE, Tanaka AC, Pomerantzeff PMA, Douay C, Charron D, Toubert A, Cunningham MW, Kalil J, et al. 2006. Mimicry in recognition of cardiac myosin peptides by heart-intralesional T cell clones from rheumatic heart disease. J Immunol. 176(9):5662–5670.
- Hemberger, M., Hanna, C. W., & Dean, W. (2020). Mechanisms of early placental development in mouse and humans. Nature Reviews. Genetics, 21(1), 27–43.
- Johnson, W. E. (2019). Origins and evolutionary consequences of ancient endogenous retroviruses. Nature Reviews. Microbiology, 17(6), 355–370.
- Komine-Aizawa, S., Takada, K., & Hayakawa, S. (2020). Placental barrier against COVID-19. Placenta, 99, 45–49.
- Maltepe, E., & Fisher, S. J. (2015). Placenta: the forgotten organ. Annual Review of Cell and Developmental Biology, 31(1), 523–552.
- Murphy K, Weaver C. 2016. Janeway’s Immunobiology. 9th ed. Boca Raton, FL: CRC Press.
- Podbilewicz, B. (2014). Virus and cell fusion mechanisms. Annual Review of Cell and Developmental Biology, 30(1), 111–139.
- Rotshenker-Olshinka, K., Volodarsky-Perel, A., Steiner, N., Rubenfeld, E., & H Dahan, M. (2020). COVID-19 pandemic effect on early pregnancy: are miscarriage rates altered, in asymptomatic women? Archives of Gynecology and Obstetrics. doi:10.1007/s00404–020–05848–0
- Rydyznski Moderbacher, C., Ramirez, S. I., Dan, J. M., Grifoni, A., Hastie, K. M., Weiskopf, D., … Crotty, S. (2020). Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell, 183(4), 996–1012.e19.
- Sivaraman, H., Er, S. Y., Choong, Y. K., Gavor, E., & Sivaraman, J. (2020). Structural basis of SARS-CoV-2- and SARS-CoV-receptor binding and small-molecule blockers as potential therapeutics. Annual Review of Pharmacology and Toxicology, 61(1). doi:10.1146/annurev-pharmtox-061220–093932
- Snow EC, Noelle RJ, Uhr JW, Vitetta ES. 1983. Activation of antigen-enriched B cells. II. Role of linked recognition in B cell proliferation to thymus-dependent antigens. J Immunol. 130(2):614–618.
- Suchman, E. (n.d.). Cytopathic effects of viruses protocols. Retrieved December 3, 2020, from Asmscience.org website: https://www.asmscience.org/docserver/fulltext/education/protocol/protocol.2875.pdf?expires=1607019012&id=id&accname=guest&checksum=3B6E28C458E5D28CFAB4499671467A46
- Turco, M. Y., & Moffett, A. (2019). Development of the human placenta. Development (Cambridge, England), 146(22), dev163428.
- Vance TDR, Lee JE. 2020. Virus and eukaryote fusogen superfamilies. Curr Biol. 30(13):R750–R754.
- Wadman, M. (2020). Why pregnant women face special risks from COVID-19. Science (New York, N.Y.). doi:10.1126/science.abe1694
- Yewdell JW, Dersh D, Fåhraeus R. 2019. Peptide channeling: The key to MHC class I immunosurveillance? Trends Cell Biol. 29(12):929–939.